-
Solutions
Solutions
Explore SciBite’s full suite of solutions to unlock the potential of your data.
-
Use Cases
Use Cases
Discover how SciBite’s powerful solutions are supporting scientists and researchers.
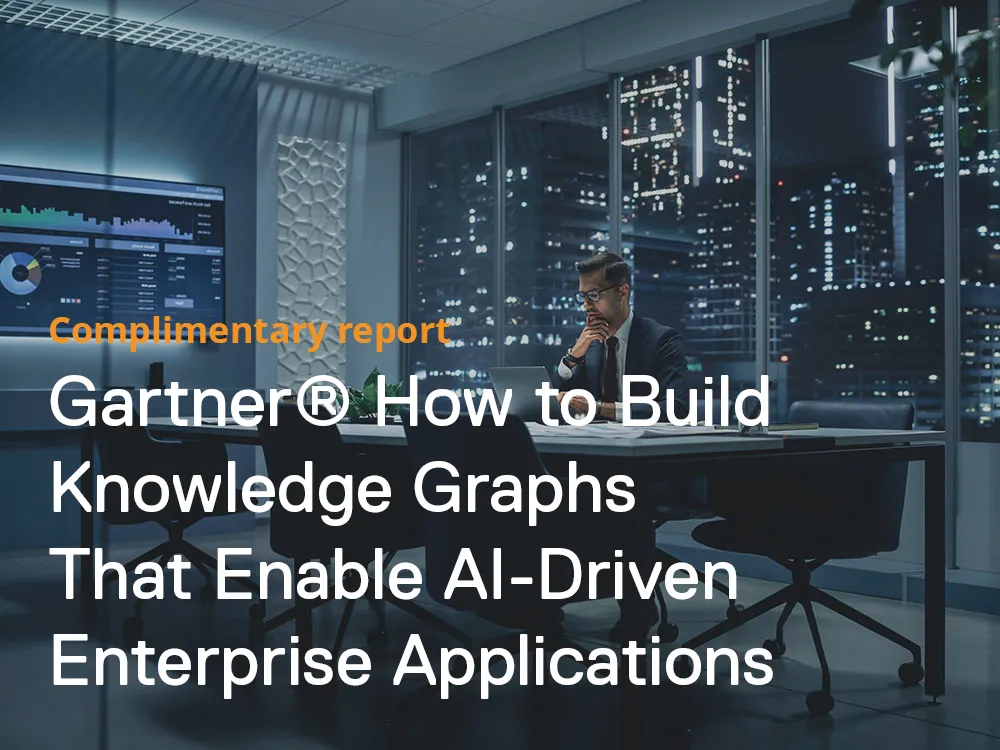
Gartner reportGartner® How to Build Knowledge Graphs That Enable AI-Driven Enterprise Applications
-
Knowledge Hub
Resources
Discover our whitepapers, spec sheets, and webinars for in-depth product knowledge.
Ctrl Alt Tech PodcastWhere technology meets curiosity. In each episode, we chat with expert guests to explore a wide range of STEM topics.
-
About
About SciBite
Explore SciBite’s full suite of solutions to unlock the potential of your data.
- Contact Us
Explore SciBite’s full suite of solutions to unlock the potential of your data.
Discover how SciBite’s powerful solutions are supporting scientists and researchers.
Gartner® How to Build Knowledge Graphs That Enable AI-Driven Enterprise Applications
Discover our whitepapers, spec sheets, and webinars for in-depth product knowledge.
Where technology meets curiosity. In each episode, we chat with expert guests to explore a wide range of STEM topics.
Explore SciBite’s full suite of solutions to unlock the potential of your data.