Text mining drug labels for genetic factors influencing efficacy and safety
Only 50-75% of patients respond beneficially to first-line drug therapy, causing unnecessary costs to healthcare providers and, more importantly, adversely impacting a patient’s quality of life. Text mining drug labels for genetic factors influencing efficacy and safety to support clinicians at the point of prescription.

Image Source: Generation Physio

A major reason for this is genetic variation affecting how individuals process a drug. Causes include differences at the drug target site or, more frequently reported, differences in the liver enzymes that metabolise the drug.
In this project we aimed to build a proof of concept tool to address the above issue. More specifically, we wanted to develop a system to meet a clinician’s requirements, nicely summarised as:
“When I am planning how to care for my patient or my population I want to know what gene tests are needed so that I prescribe the right drugs at the right dose for each individual patient.”
Solving such a challenge would be of great benefit in reducing the treatment failure rate, ultimately resulting in better quality of life for patients.
The required information is often captured on drug labels but it is a challenge for clinicians to access this information in a quick, easy and systematic way when prescribing drugs.
Consider that a GP may have less than 10 minutes to talk to a patient, come up with a diagnosis and write out a prescription: if they could access a decision support system as part of their standard IT systems to highlight known issues caused by genetic variants for any given drug, then this would enable them to propose specific genetic tests to get an idea of whether a drug will be effective for a given patient.
Furthermore, in the future, as more people have access to their personal genetic profile via services such as 23andMe, the clinician could also cross reference against this to provide more personalised health care.
The Hackathon Challenge
The work presented here is the winning entry from a challenge that was set as part of the Wellcome Genome Campus Hackathon 2018 with backing from Catapult, NHS Health Education England and the Genomics Education Programme, entitled:
“How can we combine drug and genetic data to intelligently prescribe drugs?”
Here at SciBite, we immediately saw the value in applying our text mining technology to this challenge and we signed up on the spot!
The SciBite platform was set to work to extract drugs, indications, gene/protein mentions and adverse events from prescription drug labels, and this was a key factor in the success of the tool.
Essential to the success of the project was our team of fellow Hackathon delegates from Open Targets, AstraZeneca and GSK, each offering unique skills and insights into how the challenge could be addressed. We also received valuable advice from NHS practitioners and aimed to incorporate this feedback into the platform.
The Solution: Prescription 1.5
Inspired by the overall vision of the hackathon challenge to evolve prescribing practices from “Prescription 1.0” to “Prescription 2.0”, we named our solution “Prescription 1.5” to position ourselves as a step on this evolutionary journey.
The solution developed can be described as an interruptive clinical decision support system (iCDSS), meaning that actionable information can be delivered directly to a clinician at the point of prescription. We created an integrated dataset containing genetic and drug data and a simplistic web-based user interface (WUI) to browse this data. Importantly, the WUI is served by a RESTful API, which interacts with the underlying integrated dataset and so can be embedded within existing systems.
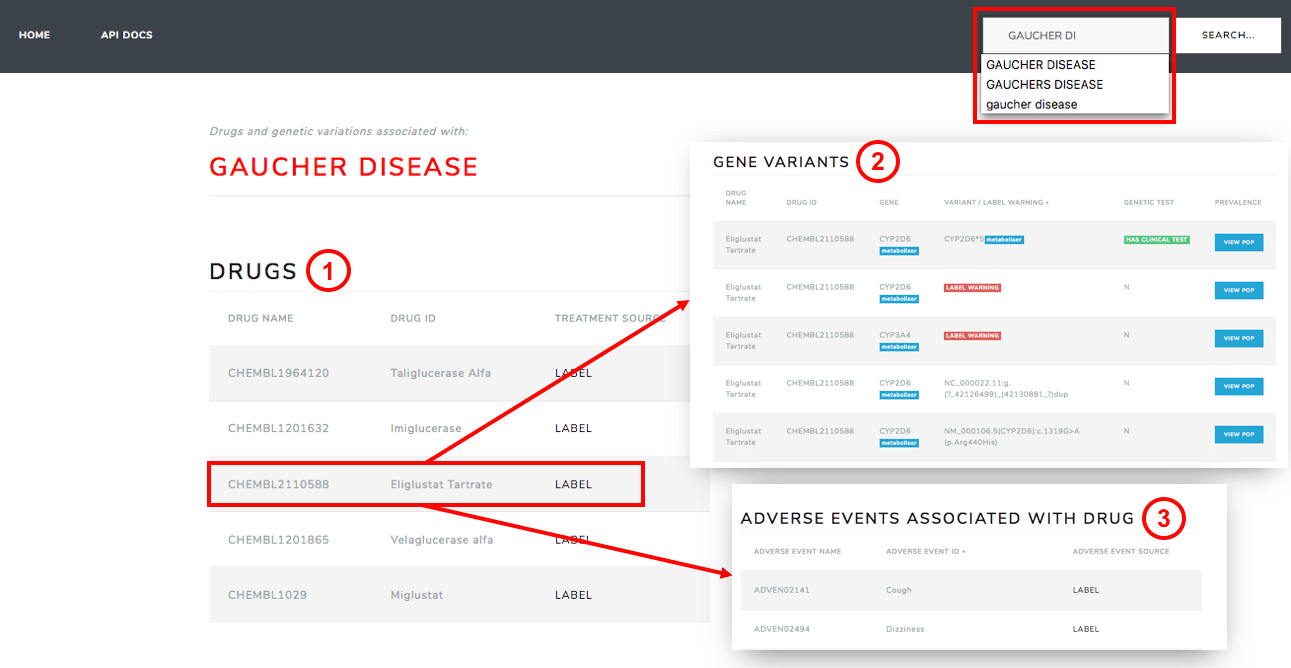
Figure 1. Web-based user interface of tool. The WUI allows a clinician to search for a particular disease (top right). A list of drug treatments are then returned (1). After selecting a particular treatment the clinician can view information regarding relevant gene variants to the treatment (2) as well as any adverse events associated with the treatment (3)
With the first iteration of the tool it is expected that a patient would visit a clinician and a diagnosis would be made. The WUI (shown in Fig.1) then allows the clinician to query the dataset with the disease that has been diagnosed. As a result all drugs that can be used to treat the query disease are returned (table 1 in Fig. 1). Once a treatment is selected, all genes/proteins mentioned in the drug label warnings section are displayed and classified as metabolisers or targets. As an advanced option, the system gives a list of any variants of these genes and the frequency of these variants within the population (table 2 in Fig. 1). If any clinical tests exist to test for the variants, these are also returned in the same table. Finally, any adverse events mentioned in the drug label are also summarised (table 3 in Fig. 1).
A RESTful API serves the WUI meaning that the functionality of the WUI can be embedded in any tool or system that may currently be in place to help a clinician during the process of prescribing treatment.
With a sleek, minimalistic and simple UI it is hoped that we have delivered a tool that has potential to greatly improve a clinicians ability to identify the best treatment for a patient based on their diagnosis and relevant genetic information. By providing an API it is hoped that the functionality of the tool can easily be embedded with other e-Prescribing tools; disrupting the current state-of-the-art.
Methods
Data integration
At the heart of the solution, we created and integrated dataset, as summarised in Fig. 2. The dataset includes drug indication data from ChEMBL, genetic test data from the Genetic Testing Registry as well as gene variant data from ClinVar. A key requirement in populating these datasets was to fill in the gaps that exist in established, manually curated datasets, such as PharmGKB, with content automatically extracted from drug labels; making use of SciBite text-mining technologies.
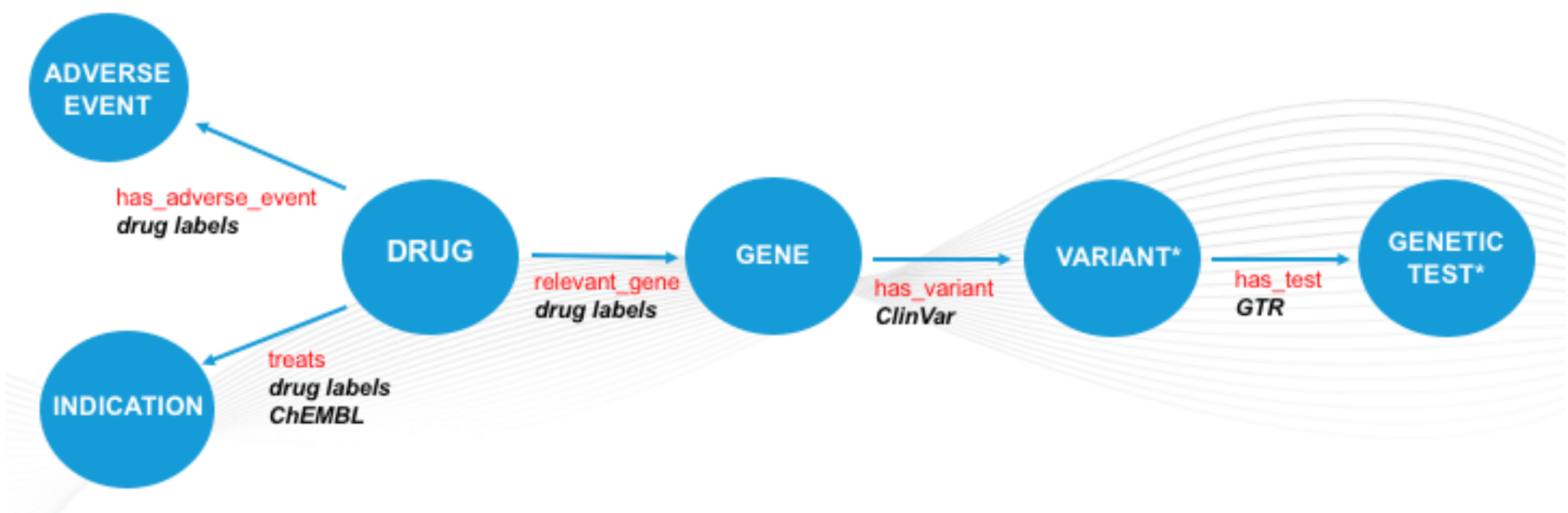
Figure 2. Metagraph of dataset. Graph representation of the data integrated during the challenge. Entities are represented as blue nodes and associations between these as blue arrows. Sources of data are captured in black. *represents entity types not populated by SciBite VOCabs
Text-Mining the Drug Labels
The SciBite platform was set to work to extract drugs, indications, gene/protein mentions and adverse events from prescription drug labels downloaded as XML from DailyMed.
The system was designed to recognise the sections of the drug label in which each entity type was mentioned. In this case, we extracted Drugs from the ‘Trade Name’ and ‘Generic Name’ sections, Indications from the ‘Indications & Usage’ section, Genes/Proteins from the ‘Warnings and Precautions’ section, and Adverse Events from the ‘Adverse Reactions’ section. There was also the possibility to mine this content for drug-drug interactions and contraindications.
Fig. 3-5 demonstrate how the technology interprets a drug label, the resulting structured, normalised data was then injected into our dataset to be assessed as part of the decision support platform.

Figure 3. Example drug label with highlighted entities. Highlighted entities Ceredegla and Eliglustat in the Trade name and Generic name sections of a drug label. Both are normalised to a single entity with the chembl identifier CHEMBL2110588, Eliglustat Tartrate
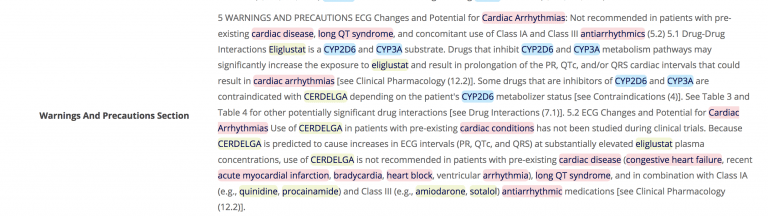
Figure 4. Example drug label with highlighted GENES. From the same drug label as Fig. 3, we can see highlighted mentions of Cytochromes P450 enzymes, CYP2D6 and CYP3A. Genetic variations in both of these are well known to affect how individuals metabolise a given drug

Figure 5. Example drug label with highlighted phrases. SciBite can also pick out phrases describing limitations of use in proximity of a Gene/Protein mention (underlined in blue)
The solution developed during this challenge shows how, with one click, a clinician may be presented with the relevant genetic and drug information required to make a better informed decision regarding a patient’s treatment with relevant genetic tests highlighted. In the future, however, the system may become more automated: with a patients sequenced genome and a diagnosis as input, a personalised treatment schedule could be returned for review by the clinician. Furthermore, by providing the tool as an API Prescription 1.5 could easily be integrated into any existing e-Prescribing software to help direct drug prescription.
As a result of rolling out this tool the time spent by clinicians to identify relevant information would be massively reduced, patients would receive personalised treatments bringing huge reductions in cost to health care providers, but most importantly, greatly improving patients quality of life!
Although there is plenty of scope for future work, the progress made within only 24 hours show cases how SciBite technologies can greatly aid in solving the challenge detailed previously; enabling data locked in a plethora of drug labels to be seamlessly integrated with other relevant data sources and systematically queried.
Find out more about SciBite’s semantic technology platform and how it can be an integral part of your business.
Related articles
-
Drug repurposing, rare diseases and semantic analytics
In this blog we cover how to look potentially reduce the cost of and speed up the repurposing pipeline.
Read -
Machine Learning and phenotype triangulation
Disease detective part 3: In our final disease detective article, we’ll take Part 2’s topic a little further and zoom in on how we can find new relationships between diseases where direct evidence is sparse.
Read
How could the SciBite semantic platform help you?
Get in touch with us to find out how we can transform your data
© Copyright © 2024 Elsevier Ltd., its licensors, and contributors. All rights are reserved, including those for text and data mining, AI training, and similar technologies.