
Matching patients to clinical trials
 |
 |
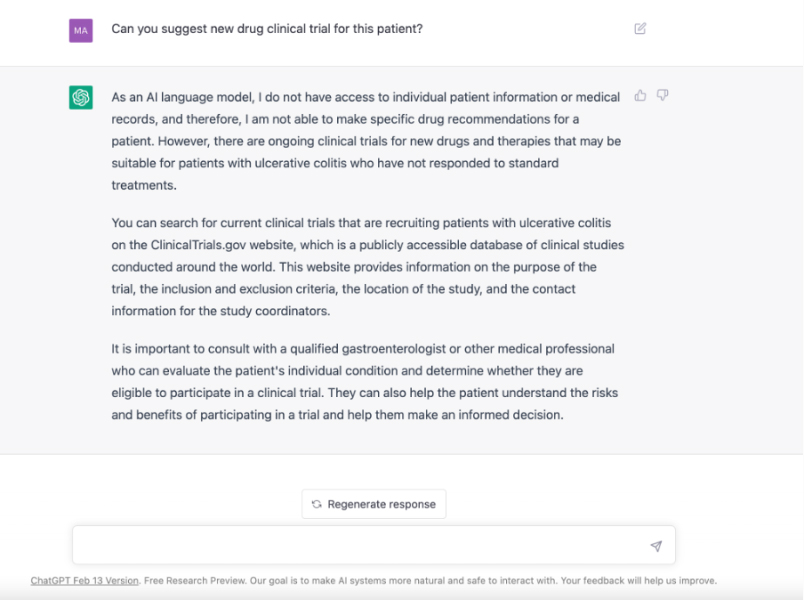
Patient X, suffering from an untreatable gastrointestinal disease, chats with a large language model for advice. GPT suggests looking at clinical trials and Patient X finds 10 active recruiting trials but is unsure which to choose. Patient X consults his doctor, who recommends a trial from a pharmaceutical company. What could go wrong?
Patient X
Patient X is watching the evening news feeling very sick from the long course of unpleasant medications his doctor prescribed for his untreatable gastrointestinal disease. But some interesting news came out about chatting with a large language model (LLM) called GPT, which claims to have passed the medical exam. Patient X heads immediately to his laptop and decides to chat with GPT about his condition.
After long chatting with GPT and finding out that all the medications it recommends were prescribed already by his doctor, he decided to ask it for new drugs. ChatGPT suggests to have a look at clinical trials at clinicaltrials.gov. Patient X searched for clinical trials and found 10 active recruiting clinical trials. However, he could not decide which one to choose from, which drug is more promising and whether he is eligible for any of them given his long history with disease and medications.
Desperate patient X tells his doctor about those trials and his doctor recommends one from a giant pharmaceutical company. The company confirmed his eligibility criteria and enrolled him into their phase 2 clinical trial. Patient X started his new medication; but, unfortunately started experiencing serious and life-threatening drug side effects.
What went wrong?
Lack of knowledge
Physicians might not be aware of all the current clinical trials in their field. And if they are, they might tend to recommend the ones from well-known and trustworthy pharmaceutical companies.
Overlooked patient medication or disease history
No physician is aware of every drug interaction and every mutated pathway or gene in the patient’s body – unless they have photographic memory. Consequently, many things can go wrong in clinical trials, If any of those mutations or interactions are missed or overlooked.
Biased AI
We are seeing an uptick in integrating AI in drug discovery and patient-drug matching, especially with the increasing numbers of LLM and high quality biomedical and biochemical data, hoping to make drug approval processes much faster and efficient. However, such AI-augmented approaches raise serious questions about:
- The provenance of LLMs training datasets and whether they are biased towards specific inclusion or exclusion criteria, drug, adverse event, or biomarker.
- Trusting openAI models with private anonymised patient medical records and matching decisions.
But, why matching patients to clinical trials is crucial?
According to FDA, any new drug should pass the following FDA processes to get on the market:
- Preclinical process, which involves new drug discovery and testing on animals
- Clinical process – Phase 1: drug safety and side effects testing on 20-80 healthy volunteers
- Clinical process – Phase 2: drug effectiveness testing on 100’s patients
- Clinical process – Phase 3: drug safety and effectiveness testing on 1000’s patients
- New drug application review (NDA), drug labelling and drug application reviewing by FDA
- Phase 4: Post-marketing safety and effectiveness assessment, which involves monitoring drug’s effects on the market
Thus, matching eligible patients to clinical process phases is considered vital for the successfullness of clinical trials and getting new and effective drugs into the market. This raises the question of how to efficiently match patients to clinical trials without overlooking or exposing any detail from any patient medical record.
Matching patients to clinical trials: R&D from SciBite
To address this question, an automated patient-matching pipeline was introduced at SciBite, using their award-winning semantic software combined with state-of-the-art AI models. The pipeline involves:
1. Matching patients and clinical trials full contexts using state-of-the-art AI models (full context matching). To achieve such matching, network analysis and centrality metrics were performed on patient-trial knowledge graphs (KG) to rank and recommend the most eligible clinical trials for each patient, using AI-generated matching scores from models, such as GPT3, BioGPT and BioBERT as well as in-house pretrained AI models (Nassar, 2109).
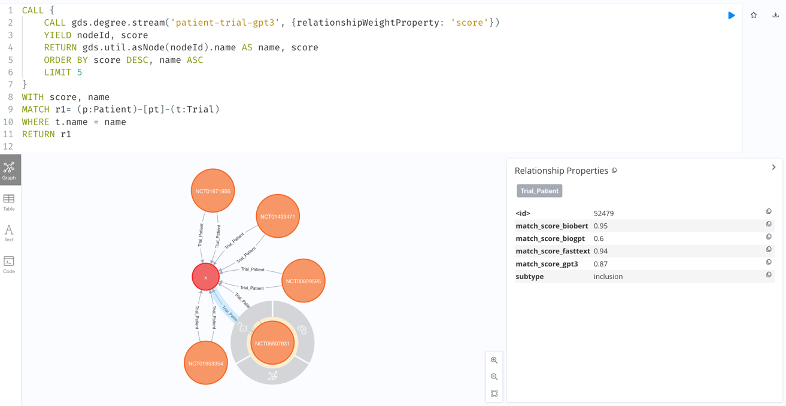
Figure 1: Screenshot of patient-trial knowledge graph showing the top 5 clinical trials for patient X, using network analysis on GPT3-generated matching score.
2. Matching patients and clinical trials entities (entity matching). Matching patients entities with clinical trials inclusion and exclusion entities can show directly if the patient is eligible or not. To match both entities, SciBite award-winning software TERMite is deployed with its high quality core and custom vocabularies in identifying and normalising most, if not all, patients and clinical trials entities, such as diseases, drugs, genes, pathways, metabolites, clinical and laboratory results along with their synonyms.
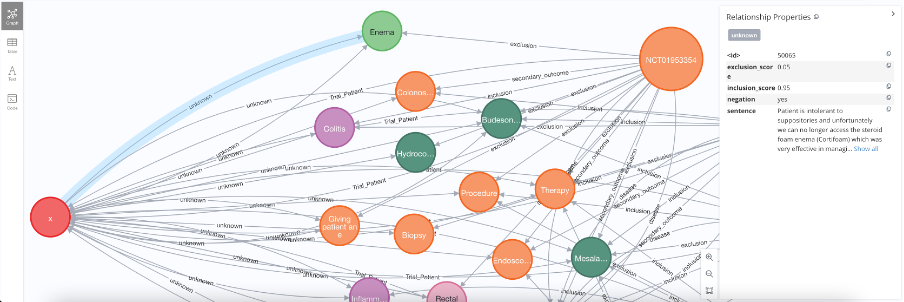
Figure 2: Screenshot of patient-trial knowledge graph showing TERMITE identifying common entities between patients medical records and clinical trials inclusion and exclusion criteria
3. Matching patients detailed contexts to those from trials inclusion or exclusion criteria (entity context matching). Whereas 1 and 2 can shortlist patients’ eligible clinical trials and match patients entities to inclusion and exclusion entities, patients detailed medication and disease history can be quite confusing and challenging, especially with drug forms, durations, intolerance, and dosage. To decipher such confusion, a cocktail of natural language processing and AI approaches were further integrated, using a) SciBite TExpress software that recognises medications dosage, disease stage scores, laboratory test results patterns and negations and b) in-house trained transformers models that generate inclusion/exclusion probabilities for patients medical records contexts and c) open access named-entity-recognition (NER) models, such as EMERALD (European Bioinformatics Institute | EMBL-EBI) host state and treatment models, in identifying patients health and disease states as well as treatments (Nassar et al., 2022).
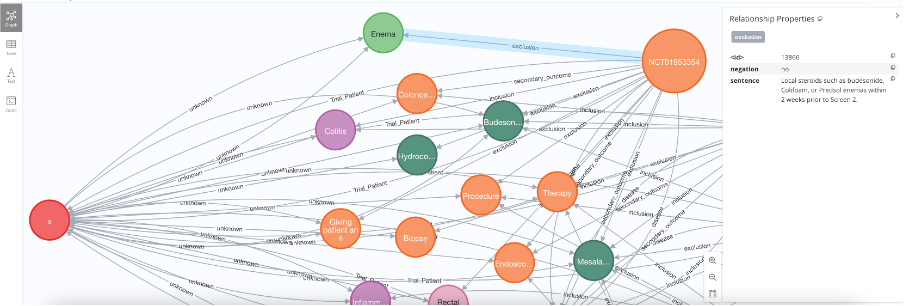
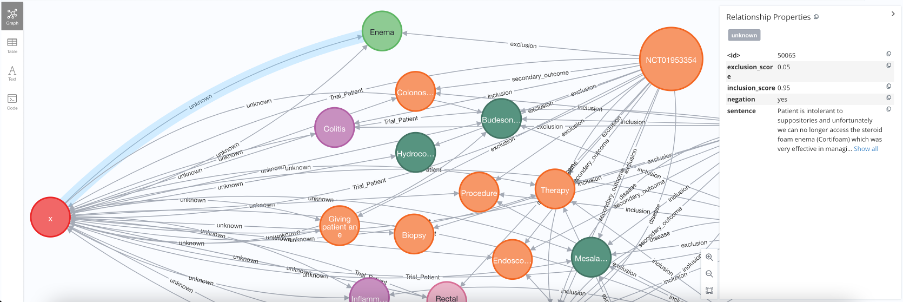
Figure 3: Screenshot of patient-trial knowledge graph showing matched entities between patients and clinical trials. Using matching score and matching entities, clinicians can exclude patients, if any of their matched entities are linked to exclusion trial edges (e.g. Enema). However, reading patient X entity contexts can reverse such decision. For example, reading Enema context reveals that patient X was intolerant to Enema, rendering him an eligible rather than uneligible candidate. Such confusion was further deciphered by identifying negated entities and generating inclusion/exclusion scores from fine-tuned transformers models.
In conclusion
In conclusion, matching patients to clinical trials is a very challenging task and crucial for new drug discovery and development. And, AI-augmented approaches showed indeed significant potentials for drug discovery and matching, especially during pandemics, when quick vaccine discovery and development is a must. However, we need to be cautious, unbiased and keep professionals in the loop when making life saving decisions. And, most importantly, not to do those decisions based on models chatting.
About Maaly Nassar
Senior Data Scientist, SciBite
Maaly joined SciBite in 2022 as a senior data scientist. With a Veterinary Medicine PhD in clinical diagnosis simulations (Freie Universitaet Berlin; FUB) and a neuroscience MSc in ML and graph analytics (Humboldt Universitaet Berlin).
Maaly developed and applied semantic computing applications and AI pipelines for data integration and enrichment (EBI EuropePMC and MGnify), medical diagnosis (clinical trials FUB), knowledge and drug discovery (EBI EuropePMC and MGnify, FUB), drug repurposing (SciBite).
Other articles by Maaly
1. [Article] The neurocognitive gains of diagnostic reasoning training using simulated interactive veterinary cases. M. Nassar, Sci Rep, 2019, 9, 19878. read more.
2. [Article] A machine learning framework for discovery and enrichment of metagenomics metadata from open access publications, M. Nassar et al., GigaScience, 2022, 11, giac077, read more.
3. [Blog] Microbiome repurposing: Is it a potential therapeutic approach & how can we do it with LLMs? read more
Related articles
-
Microbiome repurposing: Is it a potential therapeutic approach & how can we do it with LLMs?


Discover the past and future of microbiome-based healing. From ancient remedies to modern AI, learn how SciBite's groundbreaking approach blends Large Language Models (LLMs) with advanced tech to unravel the potential of therapeutic microbiomes.
Read -
Revolutionizing Life Sciences: The incredible impact of AI in Life Science [Part 1]


Artificial intelligence (AI) has been applied to numerous aspects of the life sciences, from disease diagnosis to drug discovery; in the first of this two-part blog series, we outline the impact of AI in Life Science and illustrate the various success stories of AI in Life Science.
Read
How could the SciBite semantic platform help you?
Get in touch with us to find out how we can transform your data
© Copyright © 2024 Elsevier Ltd., its licensors, and contributors. All rights are reserved, including those for text and data mining, AI training, and similar technologies.

