
Exploring breast cancer biomarkers with a literature biomarker database
In this piece we'll show how natural language processing can be applied to build a searchable database of disease biomarkers, presented in the context of their corresponding scientific publications. To illustrate the power of this approach we'll focus on examples of protein biomarkers relating to Breast Cancer.

In this piece we’ll show how natural language processing can be applied to build a searchable database of disease biomarkers, presented in the context of their corresponding scientific publications. To illustrate the power of this approach we’ll focus on examples of protein biomarkers relating to breast cancer.
What is needed from a biomarker database?
A biomarker database resource needs to ideally do two things:
- Find all the known biomarkers, so you get an understanding of current knowledge.
- Identify potential novel biomarkers to aid development of new treatment and testing strategies.
To achieve this, TExpress – SciBite’s semantically aware regular expression engine – was used to create a series of rules to identify biomarker phrases in the literature. These rules were then applied to the complete MEDLINE and NIH Grants corpora and materialised in our DOCstore semantic search platform.
Public domain biomarker databases are often focused on specific disease areas or limited to particular types of biomarkers such as gene mutations. The aim here is to add value in facilitating biomarker discovery for any disease area and for multiple types of biomarker. The types of biomarker evidence we identify include:
- General biomarker mentions – genes, miRNA, protein types, biochemicals in the presence of an indication and a biomarker context keyword
- Phrases describing biomarkers measured in a range of bodily fluids
- Phrases reporting new biomarkers
- Genetic variants in disease
Using our highly detailed VOCabs, the general biomarker mention rules alone can recognise over 2 billion possible entity combinations. If we include all the captured synonyms for these entities, then this number is several orders of magnitude greater still. Learn more in our Biomarker discovery in literature use case.
General biomarker phrases

Genetic variations
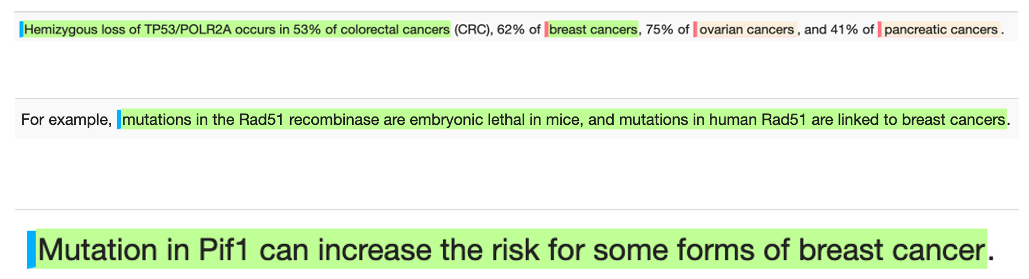
Novel biomarkers
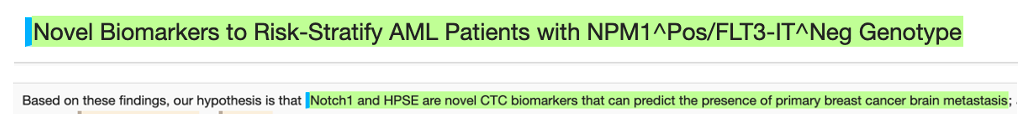
Measurements and bodily fluids

Using a literature biomarker database to identify breast cancer biomarkers
Using the above rules, the user can now quickly scan large volumes of literature to get an idea of the main biomarkers in any disease area. For the remainder of this article, we’ll focus on breast cancer with examples for identifying both well-known and more novel biomarkers.
1. What are the well-known biomarkers?
To get an overview of the most strongly associated biomarkers, through our DOCstore semantic search platform we searched for “breast cancer and “anything of type biomarker” across the past 10 years of NIH grants content. This returned over 20,000 grant submissions. We then ran these through the semantic search analytics module to get the most frequent gene/protein mentions contained within these records. Results were sorted by significance score to get the gene mentions most strongly corresponding to the breast cancer biomarker corpus weighted against the remainder of the corpus.
You can see from the chart below that erb-b2 receptor tyrosine kinase 2 (ERBB2), estrogen receptor 1 (ESR1), BRCA1 and BRCA2 come out on top. As well as providing a useful overview of the topic area, these hits confirm the validity of the method as these are well established biomarkers relating to this topic.

2. What are the novel biomarkers?
The next thing we wanted to highlight was not just the most well-known biomarkers, but to gain an understanding of what research is happening now with regards to potentially new biomarkers. To do this, we ran a sentence co-occurrence search for any gene/protein mention alongside a “novel biomarker” phrase anywhere in the text. We additionally specified that “breast cancer” must appear in the title of the grant application. The aim of this is to reduce the search space so the researcher can quickly focus in on a biomarker of interest.
Novel biomarker phrase examples for breast cancer:
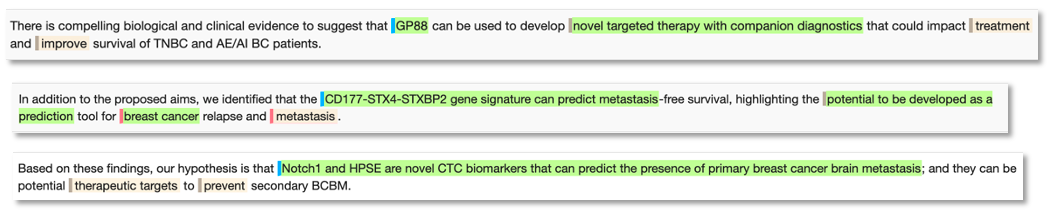
As a search tool, this provides the user with a huge biomarker resource to gain an overview of key biomarkers for any disease area. Users can manually search the content, using our semantic annotations to quickly focus on specific themes of interest. Additionally, the indexed entitles can be computationally extracted for further programmatic analysis or to populate a database resource, perhaps sitting alongside other datasets from the public domain or a company’s own content.
Searching the biomarker database: A worked example
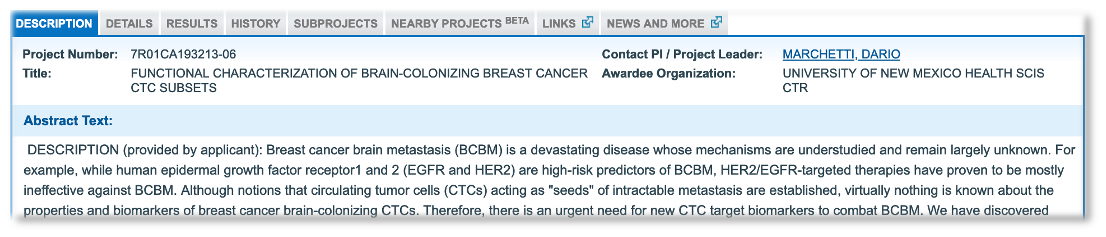
From the novel biomarker example, we picked out Notch1m for further exploration. The most recent grant application was from Dario Marchetti at the University of New Mexico, where the author makes the claim “Notch1 and HPSE can thus represent novel and specific CTC biomarkers”. In this context, CTCs are Circulating Tumor Cells involved in the spread of a cancer to other parts of the body.
Timeline analysis
To understand the context and history behind Notch1 in breast cancer, we expanded the search to Medline to explore research trends.
You can see that publications mentioning Notch1 and breast cancer (not necessarily in the biomarker context) peaked around 2015 following a steady rise in interest from 2003.
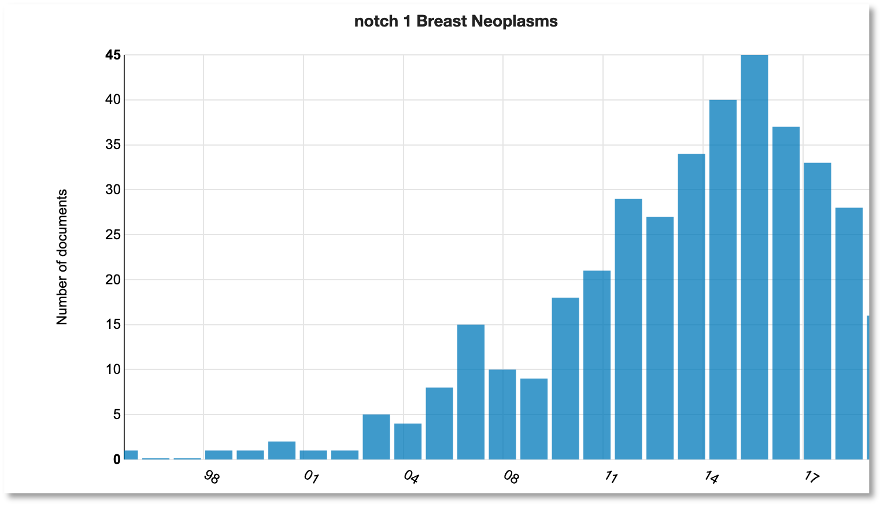
The first mention of an association between Notch1 and breast cancer was relating to mouse models dating back to 1996:
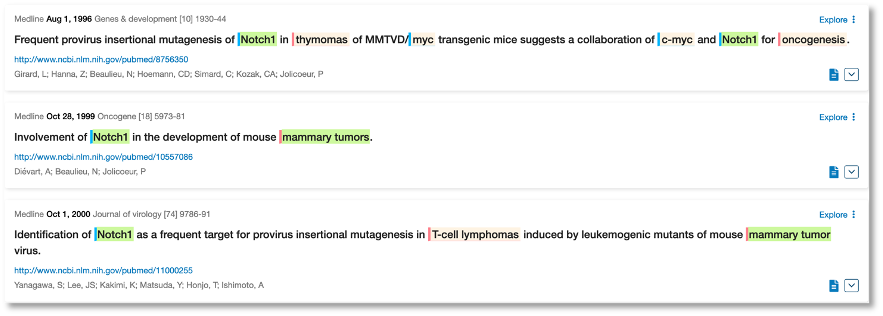
Even earlier, back to 1991, we can see fruit fly research where Notch1 (which we pick up as the synonym TAN-1) has been associated with cancer in general. We used a hierarchy search to get these
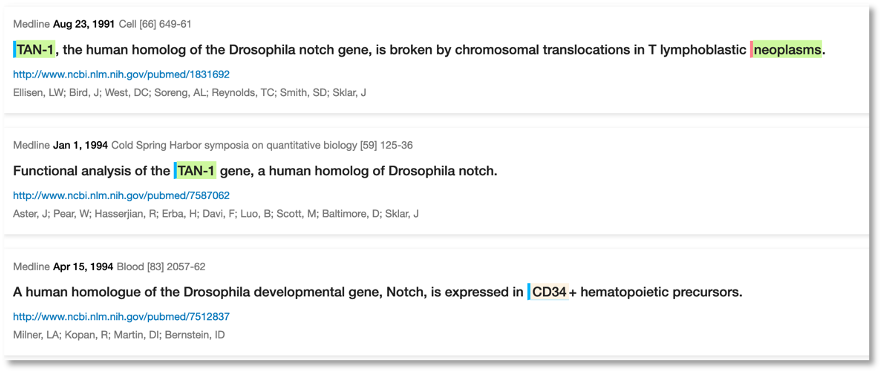
It’s interesting to see that it took around 16 years from the initial fruit fly studies to what we see today. What triggered the faster pace of publications appears to be when this was first studied in humans, for example there was a study in 2007 just before the rapid increase in publication numbers:

To the present day, we can see that the role of Notch1 in human breast cancer is well established. A sentence co-occurrence search in DOCstore give a convenient overview of the current state of research where scientists are exploring more specific mechanisms of how Notch1 is linked to breast cancer. In simpler terms, now that the link is firmly established, what is the nature of this link?
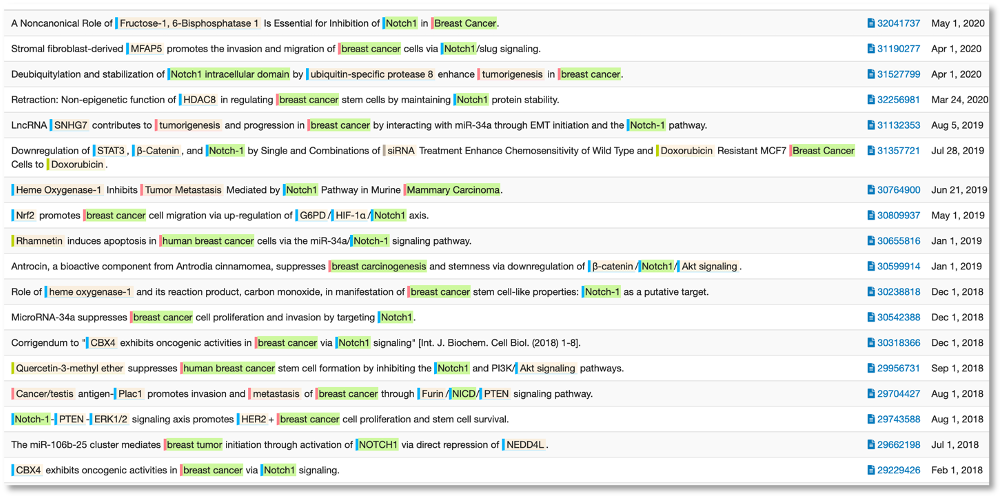
The establishment of a mechanistic link between Notch1 and breast cancer is not the same as it being a useful biomarker for the detection or prognosis of breast cancer. In fact, in amongst a number of positive papers around Notch1 as a prognostic marker, there is this:

… although reading the text more closely shows that the authors mean it is a factor of poor prognosis, rather than it is a not good a good prognostic indicator.
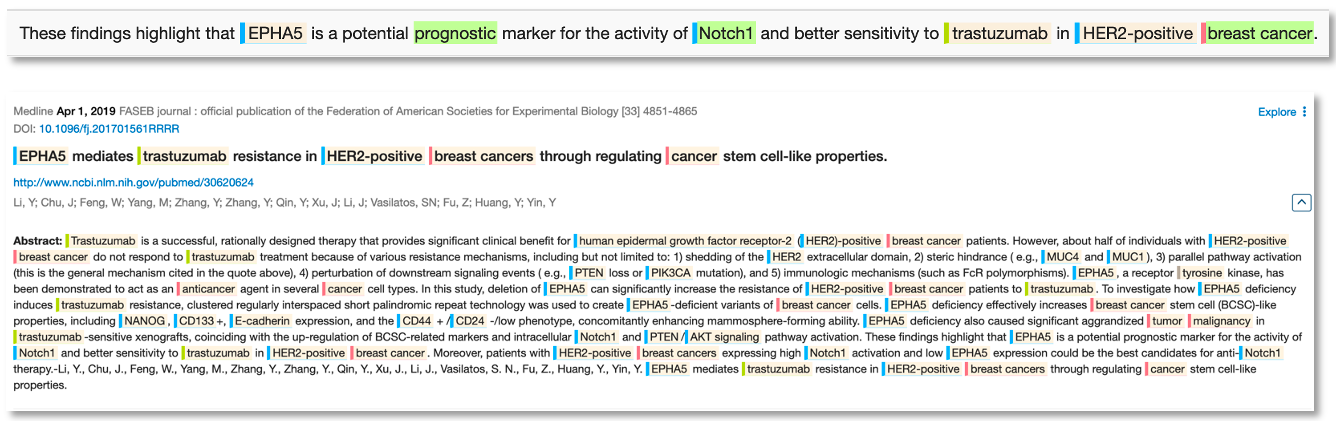
Now that scientists understand the value of Notch1 as a breast cancer biomarker, we can see from the literature that there is a new wave of research looking for biomarkers of the biomarker e.g., here we see a 2019 paper reporting EPHA5 as a marker for Notch1 activity.
What SciBite’s semantic technology has to offer
Throughout this article, we’ve demonstrated how you can use SciBite’s semantic technology to build a wide-coverage, multi-use biomarker database. The approach offers considerable advantages over an off-the-shelf biomarker database:
- As well as serving biomarkers you can utilise all the other use cases that can be addressed using our semantic text analytics technology
- You can put your own content in there, so you’re not limited to what you get with an off-the-shelf database
- You can tweak the rules and add new ones which are nice to be able to tailor it to your needs
With a simple set of rules, built on a foundation of strong ontology content designed for named entity recognition, we can rapidly construct powerful searches to support biomarker R&D. We showed how for any given disease search over a large corpus of literature, the user can quickly see the top biomarkers for that disease area, and can also focus on more novel areas of research that may be of greater interest. By investigating the publication timelines, you can follow the journey that the research took from early insect models all the way to a better understanding of specific disease mechanisms in humans.
DOCstore’s semantic search features that made this possible were:
- Named phrase annotation (based on SciBite’s BiomarkerFinder rules)
- Searching by type of entity (e.g., anything of Type Gene/Protein)
- Hierarchy search (e.g., search for any indication which is a cancer)
- Significant terms scoring (e.g., the most significant Gene/Protein mentions from my biomarker search)
- Same sentence co-occurrence searches (e.g., show me sentences that mention a novel biomarker phrase alongside a Gene/Protein mention)
- Federated and consistent search over multiple sources
To learn more about how SciBite’s semantic text analytics technology can support biomarker discovery in literature search, read our use case or get in touch with the team.
Related articles
-
Annotation of the Covid-19 open research dataset for the scientific research community
In this blog find out how the SciBite team has responded to the tech community call to arms from The White House after they released an Open Research Dataset (CORD-19), with the hope to help uncover insights and answer high-priority scientific questions related to Covid-19.
Read -
Using Enterprise Search to unlock the wealth of R&D data
Learn more in this blog post about our partnership with Sinequa and how our technologies work together to provide a winning combination of Cognitive Search and powerful Life Sciences Semantics, as we explain how enterprise search platforms are enabling Pharmaceutical companies to become more information-driven.
Read
How could the SciBite semantic platform help you?
Get in touch with us to find out how we can transform your data
© Copyright © 2024 Elsevier Ltd., its licensors, and contributors. All rights are reserved, including those for text and data mining, AI training, and similar technologies.

